
There are a variety of methods available to researchers for measuring or estimating the molecular weight of macromolecules, including gel permeation chromatography (GPC), liquid chromatography (HPLC), ultra-centrifugation, mass spectroscopy (MS), static light scattering (SLS), and dynamic light scattering (DLS). Under the category of "absolute molecular weight", ultra-centrifugation, MS, and static light scattering are the only techniques listed above that will give one an absolute measurement, i.e. in the absence of calibration standards. GPC, HPLC, and dynamic light scattering will only give molecular weight estimates, by comparison of a measured property to the same property measured for a series of standards of assumed similarity.
With regard to estimating the molecular weight from a dynamic light scattering measurement, an approach similar to that used in the GPC technique can be applied. With GPC, a molecular weight vs elution time or migration distance calibration curve is used; with DLS, a molecular weight vs hydrodynamic size calibration curve is used.
As with GPC techniques, the correlation between the specific volume (1/r) of a particle and its tertiary conformation complicates the development of a universal calibration curve for DLS. However, empirical mass vs size calibration curves, each particular to a specific ‘family’ or type of macromolecule, are available for use in DLS applications – assuming of course, that some a priori information regarding molecular conformation is known. The calibration curves used for estimating molecular weight from DLS data in the DTS software for the Malvern Zetasizer Nano system are shown in Figure 1. For reference purposes, it is noted that all of the proteins in the protein family are globular; the linear polymers are Pullulans or linear polysaccharides; the branched polymers are Ficolls or densely branched polysaccharides; and the spherical polymers are starburst type dendrimers, described as spherical, with a density that increases with radial distance from the core.
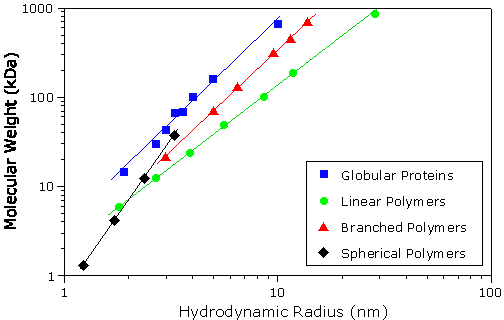
Figure 1: Molecular family dependent empirical mass vs size calibration curves for use in DLS applications for estimating molecular weight from measured hydrodynamic size.
A nice example for illustrating the value of DLS based molecular weight estimates is the evaluation of the quaternary structure of insulin. The DLS measured size distributions for human and bovine insulin at pH 2 and 7 are shown in Figure 2. At pH 2, insulin has a dimeric structure with known molecular weight of 11.4 kDa. The hydrodynamic radius at pH 2 is 1.7 nm, which is consistent with a molecular weight of 12.1 kDa using the globular protein calibration curve. At pH 7, insulin has a hexameric quaternary structure with a known molecular weight of 34.2 kDa. The radius at pH 7 is 2.7 nm, which is consistent with an estimated molecular weight of 34.1 kDa – nearly identical to the known value.


Figure 2: Size distributions for human and bovine insulin at pH 2 and 7, measured with a Malvern Instruments Zetasizer Nano system.
For additional questions or information regarding Malvern Instruments complete line of particle and materials characterization products, visit us at www.malvern.com.
